User Tools
Table of Contents
Processing Pipeline
Overview
BIAC's Processing Pipeline is a software system designed to automatically transfer data off of the scanner, run a series of automated processes on the dataset, and transfer the processed results to a directory where the user can get to it. This process depends on proper assignation of the dataset to a valid Experiment. Once assigned, data is processed according to Experiment based preferences, and the results are transferred to the appropriate Experiment directory, where users who have data access privileges can get to it.
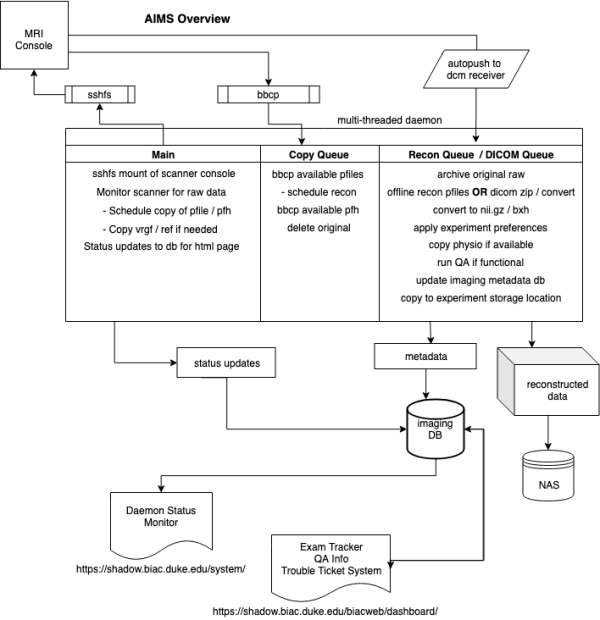
AIMS Overview
Automated Imaging Management System ( AIMS ) is a multi-threaded python daemon that runs on a desktop connected to the internal GE switch. AIMS connects to the scanner console with sshfs to monitor the hard-drive for raw k-space data and adds new files to a copy queue thread to remove them from the scanner to reconstruct and archive. AIMS launches as dicom receiver to receive automated dicom pushes from the console. Once received there is a thread monitoring the receive location for new data to schedule processing and archive. During the processing of reconstructed or dicom data images are wrapped with with BXH headers, converted to nii.gz, and copied to individual experiment storage locations. If appropriate a full BIRN QA is run for functional data, which is uploaded to our webserver as well as copied to the experiment storage location. GE physio files are also transferred if they were collected. Imaging metadata and QA numbers are added to our imaging database for use with various web services.
Backup Workflow
A nightly cron job copies locally processed and raw data from the AIMS computers to a network attached storage location. This includes all k-space data, pfile headers, nii.gz data, physio files, etc.
On a monthly basis the archives are moved to a cloud based S3 cold storage location hosted by Duke.
User Frequently Asked Questions
How does the pipeline assign a dataset to an Experiment?
For imaging data, the PatientID and ExperimentID that is entered on the scanner console is primarily used for identification. Our software takes what is entered in on the console, strips out the punctuation, and lowercases all text before before comparing against a list of valid Experiments.
The tech entered the correct Experiment ID on the scanner console, but the dataset was not properly assigned to an Experiment. What happened?
If the ExperimentID entered on the scanner does not match a list of valid Experiments, or that Experiment's storage location is full, then our processing daemon will temporarily transfer data to our Salvage.01 experiment location.
Looking for the Pipeline/Data Administration FAQ? Restricted. Request access by sending an email to Josh or Chris.
Processing Daemon History
- Processing Daemon v1.0 - Intended to process datasets per run on multiple processing servers as the datasets come over.
- Processing Daemon v1.7 - Manual intervention required for SENSE and b0 corrected data. Why? SENSE and b0 corrected recons expect the calibration run immediately preceding the run to be used. In the case that the transfer daemon transfers to mutiple nodes, there needs to be coordination among these nodes per exam, per run. We will wait until implementation of cluster processing daemon to devise a solution, as its easier to coordinate sequential run processing if there were only one datastream.
- Software ToDo List & Change Log - A constantly updated list of changes that need to be made to pipeline software in priority order and change log for systems.